Autophagy
Introduction
Autophagy is a conserved degradation pathway in eukaryotic cells that helps maintain cellular homeostasis by removing damaged organelles and misfolded proteins. While autophagy plays a critical role in normal physiological processes including cellular homeostasis, cell growth, development, and differentiation, it has also been implicated in the pathogenesis of multiple diseases including neurodegenerative disorders, cardiac myopathy, autoimmune diseases, and cancer.
Proteintech offers a wide range of products to help you with your autophagy research.
Contents: |
Autophagy pathway
Autophagy is a highly dynamic process consisting of the following three steps: (1) autophagosome formation, (2) autophagosome-lysosome fusion, and (3) degradation. It can be induced by multiple signaling pathways related to various triggers including nutrient deprivation, growth factor signaling, and cellular stress.
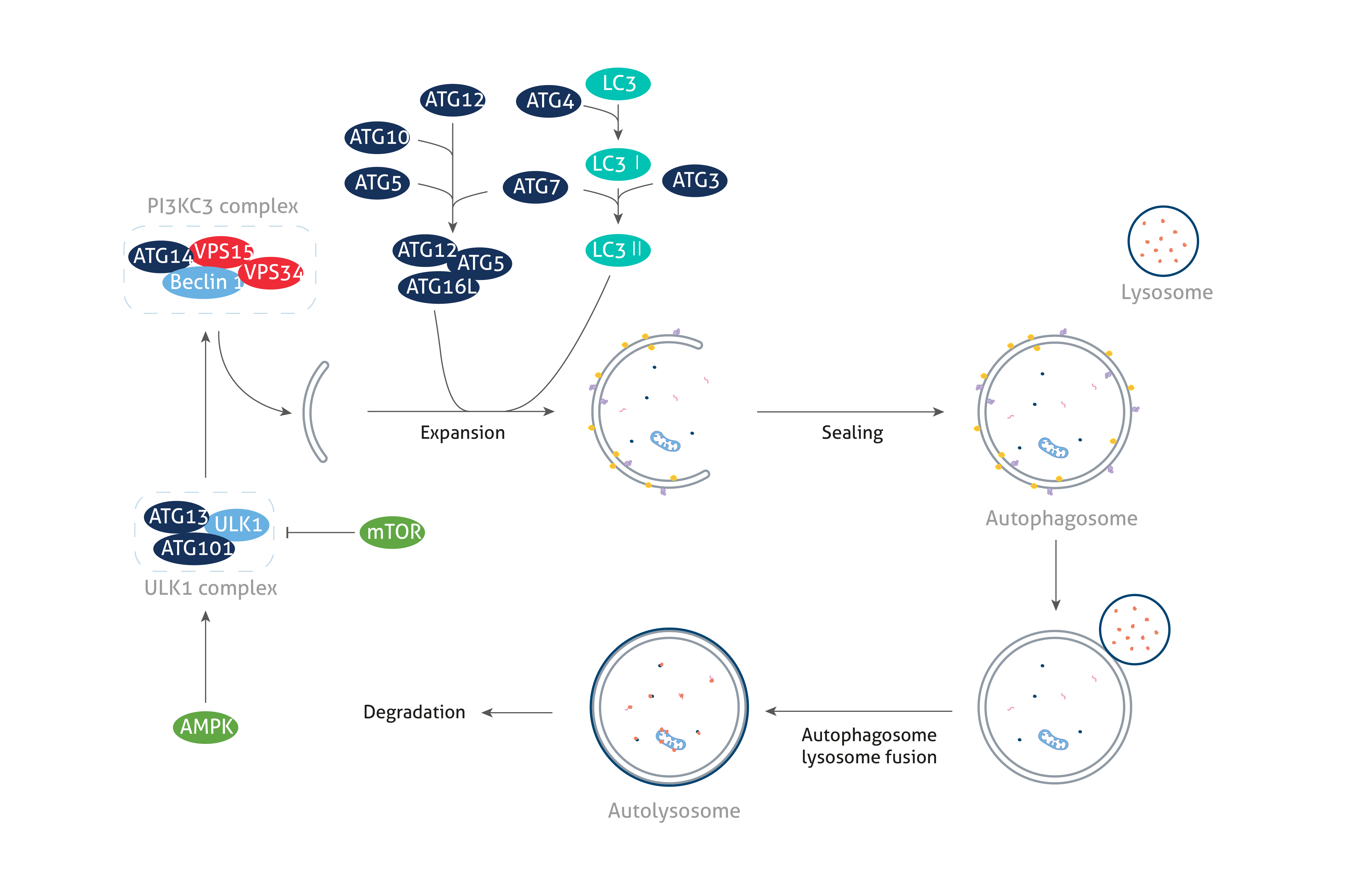
The ATG proteins
Autophagy-related (ATG) proteins are essential for the formation of autophagosomes, a critical hallmark of the autophagy pathway. The process of autophagosome formation proceeds through the steps of initiation, nucleation, elongation, closure, and ultimately fusion, each of which is regulated by various ATG proteins. Depending on their role in autophagosome biogenesis, ATG proteins can be classified into the following functional clusters: (1) the ULK1 kinase core, (2) the class III PI3K complex I, (3) the ATG2-ATG18/WIPI complex, (4) the ATG9A trafficking system, (5) the ATG5/ATG12-conjugation system, and (6) the ATG8/LC3 conjugation system.
Related products
| ULK1 | VPS15 | ATG2A | LC3A |
| ATG13 | Beclin 1 | ATG12 | ATG7 |
| RB1CC1 | ATG14 | ATG7 | ATG3 |
| ATG101 | ATG9A | ATG5 | ATG4A |
| VPS34 | WIPI1 | ATG16L1 |
Autophagic flux
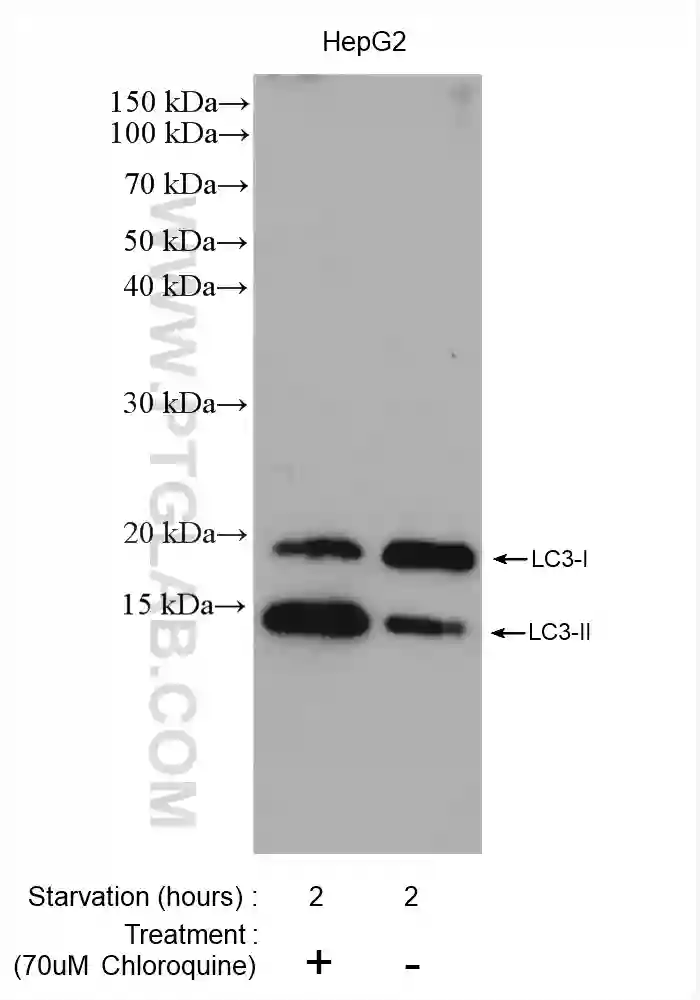
The ideal approach for measuring autophagy is to assess autophagic flux, which represents the rate of degradation of the autophagic pathway. The most widely used approach for measuring autophagic flux is to detect the processing of the autophagosomal membrane protein, LC3 by western blotting. The LC3 precursor is first cleaved by ATG4 to form LC3-I, which is then conjugated with phosphatidylethanolamine to form LC3-II. The maturation of autophagosomes into autolysosomes is followed by the degradation of inner membrane LC3-II by lysosomal proteinases. Therefore, while the induction of autophagosome formation results in an increase in LC3-II levels, the fusion of autophagosomes with lysosomes leads to a decrease in LC3-II levels.
However, simply measuring the increase in LC3-II levels may not be the optimal approach for assessing autophagy induction since decreased autophagosome and lysosome fusion can also contribute to an increase in LC3-II levels. Accurate data interpretation can be facilitated by measuring the levels of LC3-II in both the presence and absence of lysosomal inhibitors, which block the fusion of autophagosomes with lysosomes. Since treating with lysosomal inhibitors prevents autophagosomal LC3-II turnover, an increase in LC3-II levels in the presence of such inhibitors is truly indicative of increased autophagic flux. Western blot analysis of autophagy substrates such as p62/SQSTM1 is often recommended in addition to measuring LC3-II turnover for accurate assessment of autophagic flux.
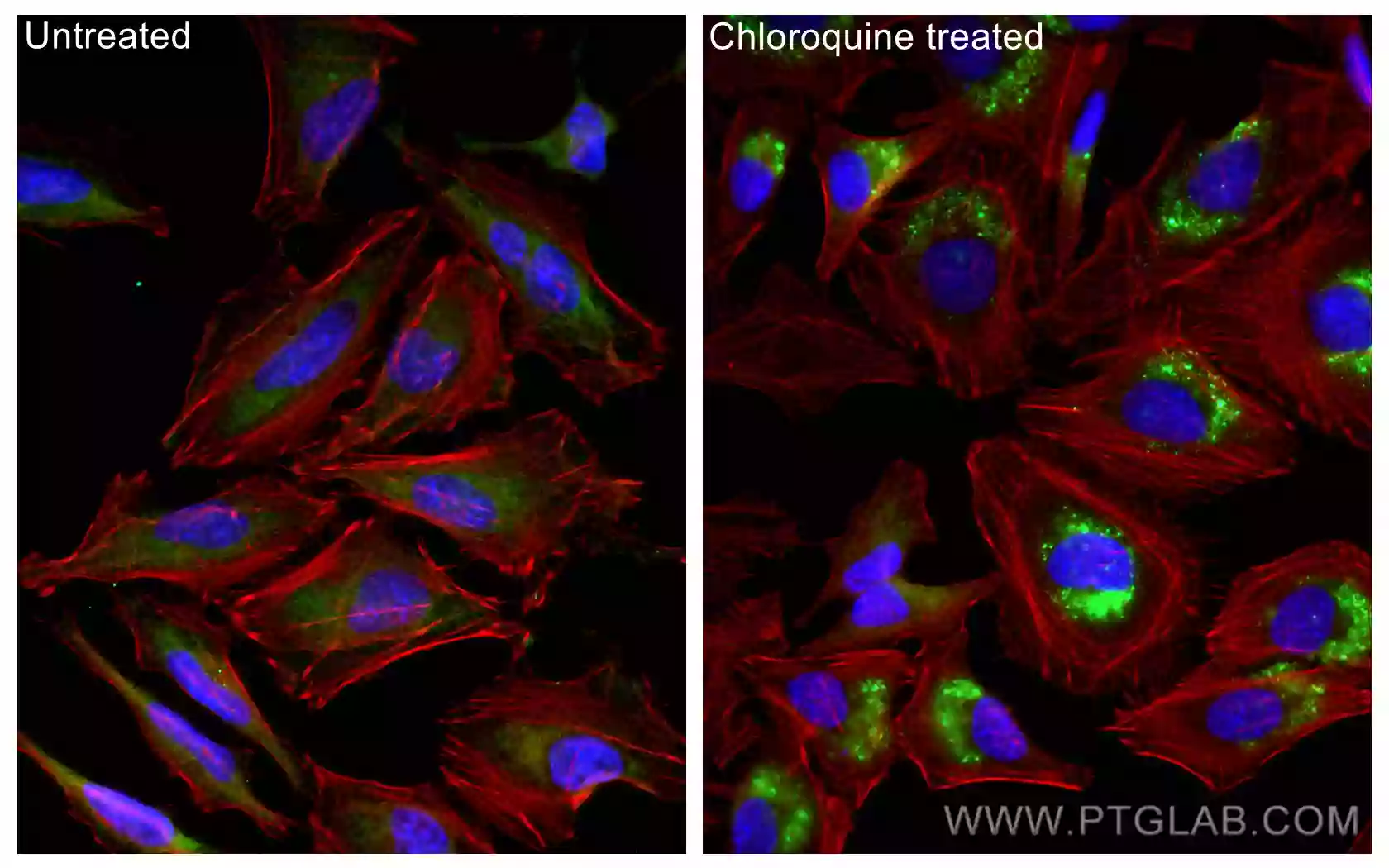
It is also possible to analyze autophagic flux using immunofluorescence by quantifying the number of LC3 puncta in the presence and absence of lysosomal inhibitors. Furthermore, the fusion of autophagosomes with lysosomes can be monitored by staining for the autophagosomal marker LC3 and the lysosomal marker, LAMP simultaneously.
Related products
| LC3 | p62 | LAMP1 |

Crosstalk between autophagy and mitochondria
Autophagy plays a critical role in the regulation of mitochondrial health by the selective degradation of damaged mitochondria as needed. This process of autophagic degradation of mitochondria, also known as mitophagy, is facilitated by a voltage-dependent kinase, PINK1, that recognizes damaged mitochondria. Mitochondrial depolarization results in PINK1 stabilization and the recruitment of Parkin onto mitochondria. Parkin ubiquitinates several mitochondrial proteins, which then leads to the recruitment of p62 and the subsequent degradation of damaged mitochondria inside autophagosomes. In addition to the removal of damaged mitochondria, mitophagy plays a role in the removal of excessive mitochondria mediated by proteins such as NIX.
While autophagy regulates mitochondrial health, mitochondria can also impact the autophagic pathway in several ways. Mitochondrial ROS, for example, is an important regulator of autophagy. Mitochondria can directly regulate autophagic flux by inducing mTOR or AMPK-mediated autophagy when mitochondrial ATP production decreases. Moreover, mitochondria have been suggested as a potential source for autophagosomal membranes during starvation-induced autophagy. Changes in the rates of mitochondrial fusion and fission, known as mitochondrial dynamics, can also influence the way cells respond to autophagy. For example, mitochondria have been shown to undergo excessive fusion during starvation as a mechanism to protect themselves against autophagic degradation.
Related products
| Fis1 | Mfn2 | Parkin |
| Mfn1 | BNIP3L | PINK1 |