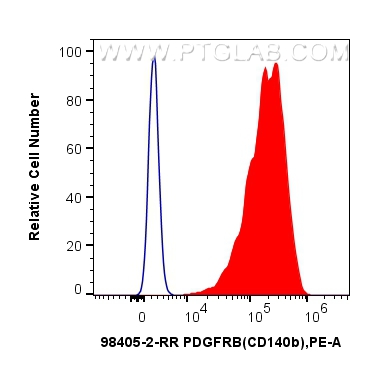
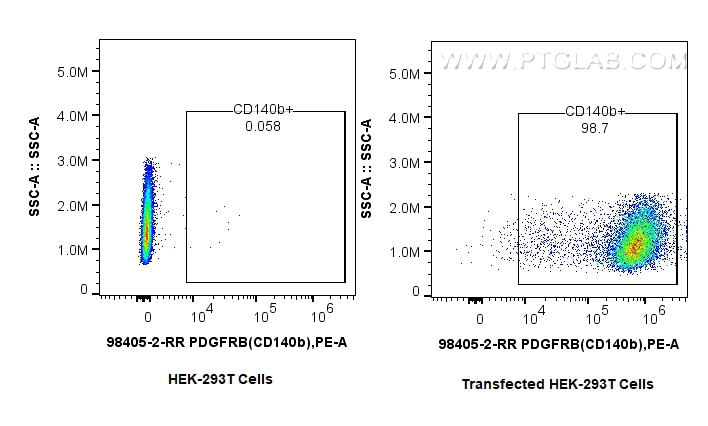
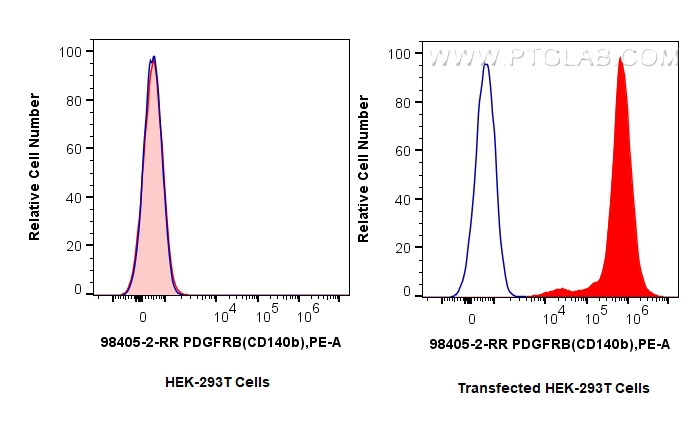
验证数据展示
经过测试的应用
| Positive FC detected in | MG-63 cells, Transfected HEK-293T cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Flow Cytometry (FC) | FC : 0.25 ug per 10^6 cells in 100 μl suspension |
| This reagent has been tested for flow cytometric analysis. It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
98405-2-RR targets PDGFR beta/CD140b in FC applications and shows reactivity with human samples.
| 经测试应用 | FC Application Description |
| 经测试反应性 | human |
| 免疫原 |
CatNo: Eg3715 Product name: recombinant human PDGFRB protein Source: mammalian cells-derived, pHZ-KIsec-C-rFc Tag: C-rFc Domain: 33-531 aa of BC032224 Sequence: LVVTPPGPELVLNVSSTFVLTCSGSAPVVWERMSQEPPQEMAKAQDGTFSSVLTLTNLTGLDTGEYFCTHNDSRGLETDERKRLYIFVPDPTVGFLPNDAEELFIFLTEITEITIPCRVTDPQLVVTLHEKKGDVALPVPYDHQRGFFGIFEDRSYICKTTIGDREVDSDAYYVYRLQVSSINVSVNAVQTVVRQGENITLMCIVIGNEVVNFEWTYPRKESGRLVEPVTDFLLDMPYHIRSILHIPSAELEDSGTYTCNVTESVNDHQDEKAINITVVESGYVRLLGEVGTLQFAELHRSRTLQVVFEAYPPPTVLWFKDNRTLGDSSAGEIALSTRNVSETRYVSELTLVRVKVAEAGHYTMRAFHEDAEVQLSFQLQINVPVRVLELSESHPDSGEQTVRCRGRGMPQPNIIWSACRDLKRCPRELPPTLLGNSSEEESQLETNVTYWEEEQEFEVVSTLRLQHVDRPLSVRCTLRNAVGQDTQEVIVVPHSLPFK 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Recombinant |
| 产品类型 | Antibody |
| 全称 | platelet-derived growth factor receptor, beta polypeptide |
| 别名 | PDGFRB, Beta platelet-derived growth factor receptor, Beta-type platelet-derived growth factor receptor, CD140 antigen-like family member B, CD140B |
| 计算分子量 | 1106 aa, 124 kDa |
| GenBank蛋白编号 | BC032224 |
| 基因名称 | PDGFR beta |
| Gene ID (NCBI) | 5159 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Protein A purification |
| UNIPROT ID | P09619 |
| 储存缓冲液 | PBS with 0.09% sodium azide, pH 7.3. |
| 储存条件 | Store at 2 - 8°C. Stable for one year after shipment. |
背景介绍
PDGFRs are receptor tyrosine kinases with an extracellular ligand-binding domain and a cytoplasmic enzymatic domain. The signaling activity of PDGFRs is tightly regulated by an autoinhibitory allosteric conformation, and requires ligand-induced receptor dimerization at the cell surface to alleviate autoinhibition and achieve physiological activity (PMID: 21664579). PDGFR beta (platelet-derived growth factor receptor- β)/CD140b is a constitutive marker expressed by ASCs in the angiogenic behavior of human retinal endothelial cells (HREs). CD140b (PDGFRβ) plays a crucial role in controlling the intrinsic proliferation (PMID: 30976657)
实验方案
| Product Specific Protocols | |
|---|---|
| FC protocol for PDGFR beta/CD140b antibody 98405-2-RR | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |