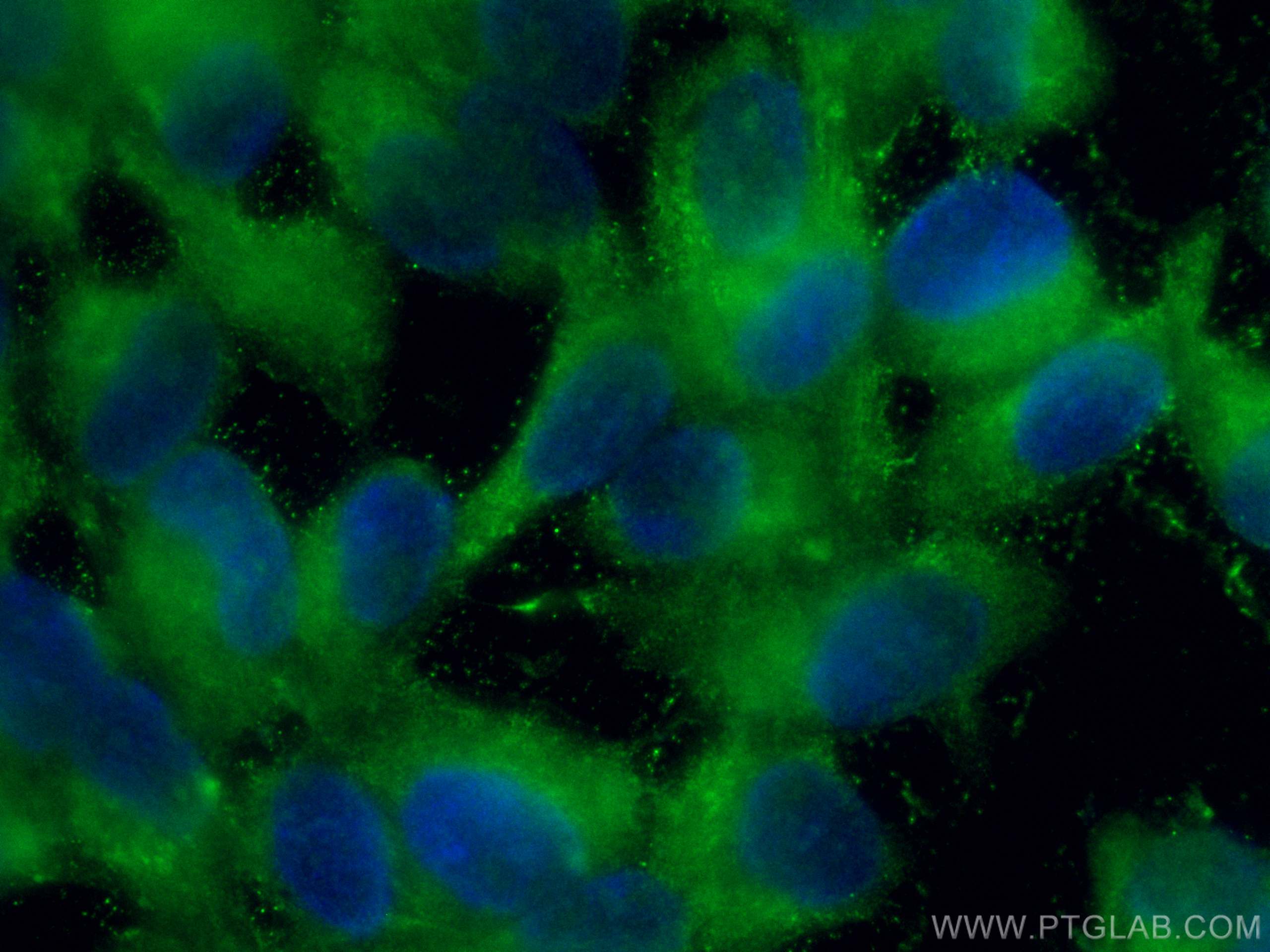
验证数据展示
经过测试的应用
| Positive IF/ICC detected in | hTERT-RPE1 cells |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Immunofluorescence (IF)/ICC | IF/ICC : 1:50-1:500 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
CL488-24430 targets RS1 in IF/ICC applications and shows reactivity with human, mouse, rat samples.
| 经测试应用 | IF/ICC Application Description |
| 经测试反应性 | human, mouse, rat |
| 免疫原 |
CatNo: Ag18885 Product name: Recombinant human RS1 protein Source: e coli.-derived, PET28a Tag: 6*His Domain: 25-224 aa of BC141638 Sequence: TEDEGEDPWYQKACKCDCQGGPNALWSAGATSLDCIPECPYHKPLGFESGEVTPDQITCSNPEQYVGWYSSWTANKARLNSQGFGCAWLSKFQDSSQWLQIDLKEIKVISGILTQGRCDIDEWMTKYSVQYRTDERLNWIYYKDQTGNNRVFYGNSDRTSTVQNLLRPPIISRFIRLIPLGWHVRIAIRMELLECVSKCA 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Polyclonal |
| 产品类型 | Antibody |
| 全称 | retinoschisin 1 |
| 别名 | Retinoschisin, retinoschisin 1, RS, RS1, XLRS1 |
| 计算分子量 | 26 kDa |
| 观测分子量 | 26 kDa |
| GenBank蛋白编号 | BC141638 |
| 基因名称 | RS1 |
| Gene ID (NCBI) | 6247 |
| RRID | AB_3084081 |
| 偶联类型 | CoraLite® Plus 488 Fluorescent Dye |
| 最大激发/发射波长 | 493 nm / 522 nm |
| 形式 | Liquid |
| 纯化方式 | Antigen affinity purification |
| UNIPROT ID | O15537 |
| 储存缓冲液 | PBS with 50% glycerol, 0.05% Proclin300, 0.5% BSA, pH 7.3. |
| 储存条件 | Store at -20°C. Avoid exposure to light. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
RS1 is also named as XLRS1. RS1 can bind negatively charged membrane lipids, such as phosphatidylserine and phosphoinositides. RS1 is required for normal structure and function of the retina (PMID:19093009). It may play a role in cell-cell adhesion processes in the retina (PMID:27114531). RS1 is high expression in the retina (at protein level) (PMID:10915776, PMID:9326935). It is detected in the inner segment of the photoreceptors, the inner nuclear layer, the inner plexiform layer and the ganglion cell layer. At the macula, it is expressed in both the outer and inner nuclear layers and in the inner plexiform layer (PMID:10915776, PMID:10915776). And RS1 is undetectable in the inner plexiform layers and the inner nuclear layer (PMID:10915776)
实验方案
| Product Specific Protocols | |
|---|---|
| IF protocol for CL Plus 488 RS1 antibody CL488-24430 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |