验证数据展示
经过测试的应用
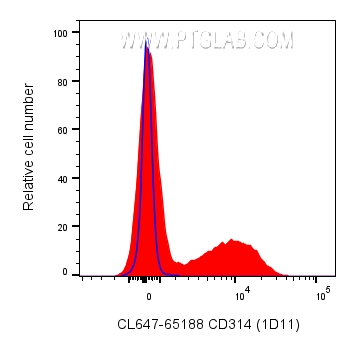
| Positive FC detected in | human PBMCs |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Flow Cytometry (FC) | FC : 5 ul per 10^6 cells in 100 μl suspension |
| This reagent has been pre-titrated and tested for flow cytometric analysis. The suggested use of this reagent is 5 μl per 10^6 cells in a 100 µl suspension or 5 μl per 100 µl of whole blood. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
CL647-65188 targets CD314/NKG2D in FC applications and shows reactivity with Human samples.
| 经测试应用 | FC Application Description |
| 经测试反应性 | Human |
| 免疫原 |
NKL cell line 种属同源性预测 |
| 宿主/亚型 | Mouse / IgG1, kappa |
| 抗体类别 | Monoclonal |
| 产品类型 | Antibody |
| 全称 | killer cell lectin-like receptor subfamily K, member 1 |
| 别名 | CD314, D12S2489E, FLJ17759, FLJ75772, KLR, KLRK1, NK cell receptor D, NKG2 D, NKG2 D activating NK receptor, NKG2D |
| 计算分子量 | 25 kDa |
| GenBank蛋白编号 | BC039836 |
| 基因名称 | KLRK1 |
| Gene ID (NCBI) | 22914 |
| RRID | AB_3673576 |
| 偶联类型 | CoraLite® Plus 647 Fluorescent Dye |
| 最大激发/发射波长 | 654 nm / 674 nm |
| 形式 | Liquid |
| 纯化方式 | Affinity purification |
| UNIPROT ID | P26718 |
| 储存缓冲液 | PBS with 0.09% sodium azide and 0.5% BSA, pH 7.3. |
| 储存条件 | Store at 2-8°C. Avoid exposure to light. Stable for one year after shipment. |
背景介绍
CD314, also known as NKG2D or Killer cell lectin-like receptor subfamily K member 1 (KLRK1), is a type II lectin-like transmembrane stimulatory receptor (PMID: 8436421). In humans, it is expressed on NK cells, gamma delta T cells, and CD8+ alpha beta T cells (PMID: 10426993). Various families of cell surface ligands have been identified, including the MICA/MICB and ULBP proteins (PMID: 12150888). CD314 is involved in both innate and adaptive immunities, and the NKG2D/NKG2DL pathway involves multiple effector cell types for controlling tumor progression (PMID: 31720075).
实验方案
| Product Specific Protocols | |
|---|---|
| FC protocol for CL Plus 647 CD314/NKG2D antibody CL647-65188 | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |