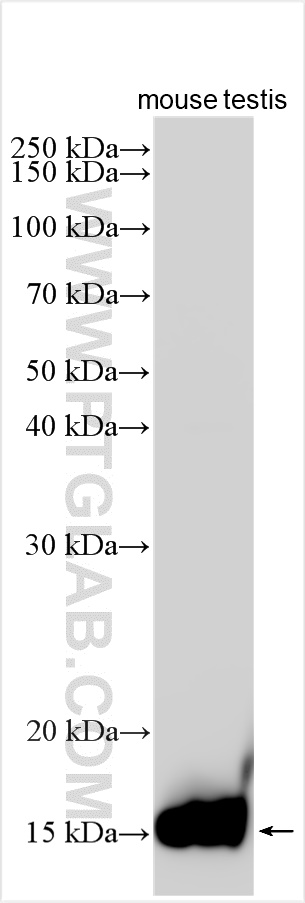
验证数据展示
经过测试的应用
| Positive WB detected in | mouse testis tissue |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Western Blot (WB) | WB : 1:1000-1:6000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
31048-1-AP targets Histone H2B in WB, ELISA applications and shows reactivity with Human, mouse samples.
| 经测试应用 | WB, ELISA Application Description |
| 经测试反应性 | Human, mouse |
| 免疫原 |
CatNo: Ag33975 Product name: Recombinant human HIST1H2BA protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 1-127 aa of BC066242 Sequence: MPEVSSKGATISKKGFKKAVVKTQKKEGKKRKRTRKESYSIYIYKVLKQVHPDTGISSKAMSIMNSFVTDIFERIASEASRLAHYSKRSTISSREIQTAVRLLLPGELAKHAVSEGTKAVTKYTSSK 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Polyclonal |
| 产品类型 | Antibody |
| 全称 | histone cluster 1, H2ba |
| 别名 | bA317E16.3, H2BFU, HIST1H2BA, histone cluster 1, H2ba, Histone H2B type 1 A, Histone H2B, testis, STBP, Testis specific histone H2B, TSH2B |
| 观测分子量 | 15 kDa |
| GenBank蛋白编号 | BC066242 |
| 基因名称 | Histone H2B |
| Gene ID (NCBI) | 255626 |
| RRID | AB_3669832 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Antigen affinity purification |
| UNIPROT ID | Q96A08 |
| 储存缓冲液 | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| 储存条件 | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
Histones are the basic nuclear proteins responsible for the nucleosome structure of the chromosomal fiber. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures.Histone H2BC1 is testis specific.
实验方案
| Product Specific Protocols | |
|---|---|
| WB protocol for Histone H2B antibody 31048-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |