验证数据展示
经过测试的应用
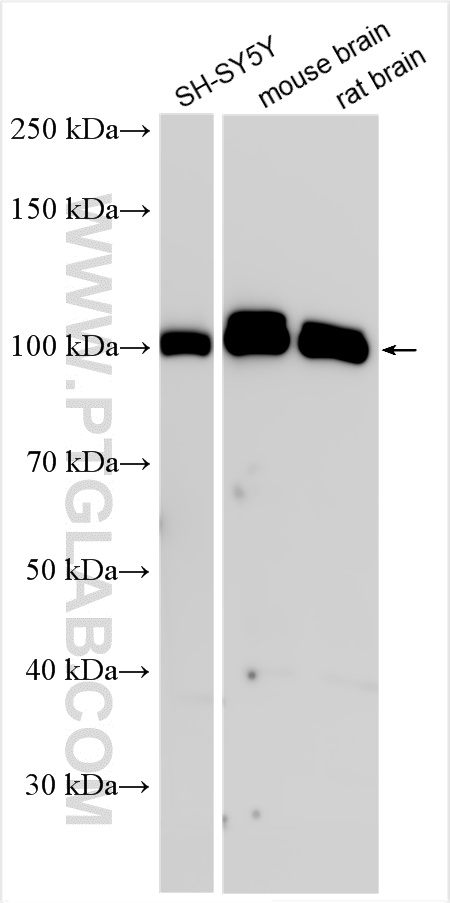
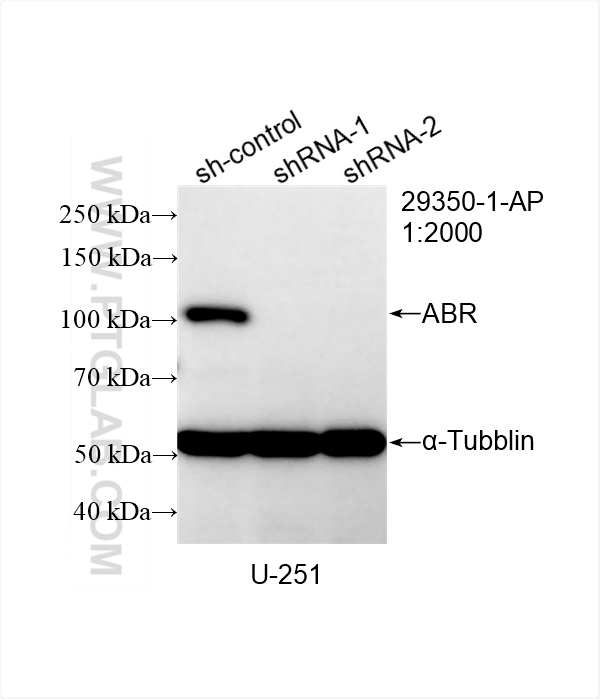
| Positive WB detected in | SH-SY5Y cells, U-251 cells, mouse brain tissue, rat brain tissue |
| Positive IHC detected in | rat brain tissue Note: suggested antigen retrieval with TE buffer pH 9.0; (*) Alternatively, antigen retrieval may be performed with citrate buffer pH 6.0 |
推荐稀释比
| 应用 | 推荐稀释比 |
|---|---|
| Western Blot (WB) | WB : 1:1000-1:4000 |
| Immunohistochemistry (IHC) | IHC : 1:500-1:2000 |
| It is recommended that this reagent should be titrated in each testing system to obtain optimal results. | |
| Sample-dependent, Check data in validation data gallery. | |
产品信息
29350-1-AP targets ABR in WB, IHC, ELISA applications and shows reactivity with human, mouse, rat samples.
| 经测试应用 | WB, IHC, ELISA Application Description |
| 经测试反应性 | human, mouse, rat |
| 免疫原 |
CatNo: Ag29522 Product name: Recombinant human ABR protein Source: e coli.-derived, PGEX-4T Tag: GST Domain: 1-87 aa of NM_021962 Sequence: MEPLSHRGLPRLSWIDTLYSNFSYGTDEYDGEGNEEQKGPPEGSETMPYIDESPTMSPQLSARSQGGGDGVSPTPPEGLAPGVEAGK 种属同源性预测 |
| 宿主/亚型 | Rabbit / IgG |
| 抗体类别 | Polyclonal |
| 产品类型 | Antibody |
| 全称 | active BCR-related gene |
| 别名 | active BCR related gene, MDB |
| 计算分子量 | 98 kDa |
| 观测分子量 | 97-100 kDa |
| GenBank蛋白编号 | NM_021962 |
| 基因名称 | ABR |
| Gene ID (NCBI) | 29 |
| RRID | AB_3086122 |
| 偶联类型 | Unconjugated |
| 形式 | Liquid |
| 纯化方式 | Antigen affinity purification |
| UNIPROT ID | Q12979 |
| 储存缓冲液 | PBS with 0.02% sodium azide and 50% glycerol, pH 7.3. |
| 储存条件 | Store at -20°C. Stable for one year after shipment. Aliquoting is unnecessary for -20oC storage. |
背景介绍
ABR (Active breakpoint cluster region-related protein) is the only protein known in humans and mice to share high homology with BCR (68% amino acid identity). BCR(Breakpoint cluster region) gene was originally identified due to its involvement in a specific chromosomal translocation that causes the development of chronic myeloid leukemia and a subset of acute lymphoblastic leukemia. The C-terminus is a GTPase-activating protein domain which stimulates GTP hydrolysis by RAC1, RAC2 and CDC42. (PMID: 17116687, PMID: 37507586)
实验方案
| Product Specific Protocols | |
|---|---|
| IHC protocol for ABR antibody 29350-1-AP | Download protocol |
| WB protocol for ABR antibody 29350-1-AP | Download protocol |
| Standard Protocols | |
|---|---|
| Click here to view our Standard Protocols |