To dye or not to dye? Understanding the different viability dyes available for flow cytometry
Dead cell exclusion in flow cytometry is crucial for reliable and accurate results. This blog will help you understand which reagents are best for your experiment, providing a comprehensive overview of the popular products available to help you find "another way to dye".
When performing a flow cytometry experiment, it is critical to exclude dead cells in your sample to generate reliable, accurate results. Dead cells can interfere with analysis due to i) non-specific binding to antibodies, ii) having a higher degree of autofluorescence than living cells, and iii) cell clumping due to the release of DNA. As expected, this can cause the resulting data to be inaccurate, especially when observing low-expression antigens, and can compromise cell-sorting.
There are many different reagents available to perform this exclusion, and they each have their different uses, advantages, and disadvantages. This blog aims to educate the user on how they work and in which situations they are best suited.
DNA binding dyes
During cell death, the plasma and nuclear membranes break down and become permeable. As such, dyes like Propidium Iodide (PI), 7-AAD, and DAPI can enter the cell and bind to DNA. Protocols for using these are relatively straightforward, and typically involve a short incubation time for the dye right before running on the cytometer. Be aware that cells are continuously dying in your sample, so ensure that dye is added at a consistent time before analysis between samples for accurate comparison. The drawback of DNA binding dyes is that they take advantage of dead cells’ membrane permeability, so they cannot distinguish between living and dead cells that have been fixed.
Below is a table summarizing the different DNA binding dyes available:
|
PI (Propidium iodide) |
Excitation: 488 nm Emission: 617 nm |
Binds to double stranded DNA by intercalating base pairs. Can be used with other fluorophores excited by 488 (e.g., FITC or CoraLite488) due to large Stokes shift. Protocol example: PMID 27371595 |
|
7-AAD (7-amino actinomycin D) |
Excitation: 488 nm Emission: 647 nm |
Binds to double stranded DNA by intercalating base pairs in G-C rich regions. Can be used with other fluorophores excited by 488 (e.g., FITC or CoraLite488) due to large Stokes shift. Protocol example: PMID 7512891 |
|
DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride) |
Excitation: 358 nm Emission: 461 |
Can be excited by a violet laser, e.g., 405 nm. DAPI binds to double stranded DNA at A-T rich regions. Also binds to RNA, emitting at 500 nm and at lower intensity. Protocol example: PMID 27037070 |
Fixable viability dyes
As discussed above, DNA binding dyes can only discriminate dead from live cells in unfixed samples. However, there are many methods available that can identify dead cells in fixed samples. These are fixable dyes that react with protein amine groups in both dead and live cells, yet due to the compromised cell membrane in dead cells the dye can bind to more proteins inside the cell (Figure 1). This results in a much higher fluorescence (roughly 50x greater) in dead cells, allowing live cells to be easily identified. Fixable viable dyes, aptly named Zombie Dyes™ (Bio-Legend), Ghost Dyes™ (Tonbo Biosciences) etc., are popular due to their ease of use and availability in many colors to suit different panel designs. Proteintech has recently launched a range of fixable viability dyes, Phantom Dyes, available in 7 different excitation/emission ranges (see below). Phantom Dyes are covalently attached to cellular proteins, thus remain bound and viable following fixation with formaldehyde and alcohol reagents. These dyes can also be used in unfixed live cells, making them an excellent addition for every flow cytometry panel, and can also be used as a compensation control when paired with amine-reactive beads. For a detailed protocol, see PMID 20578108.
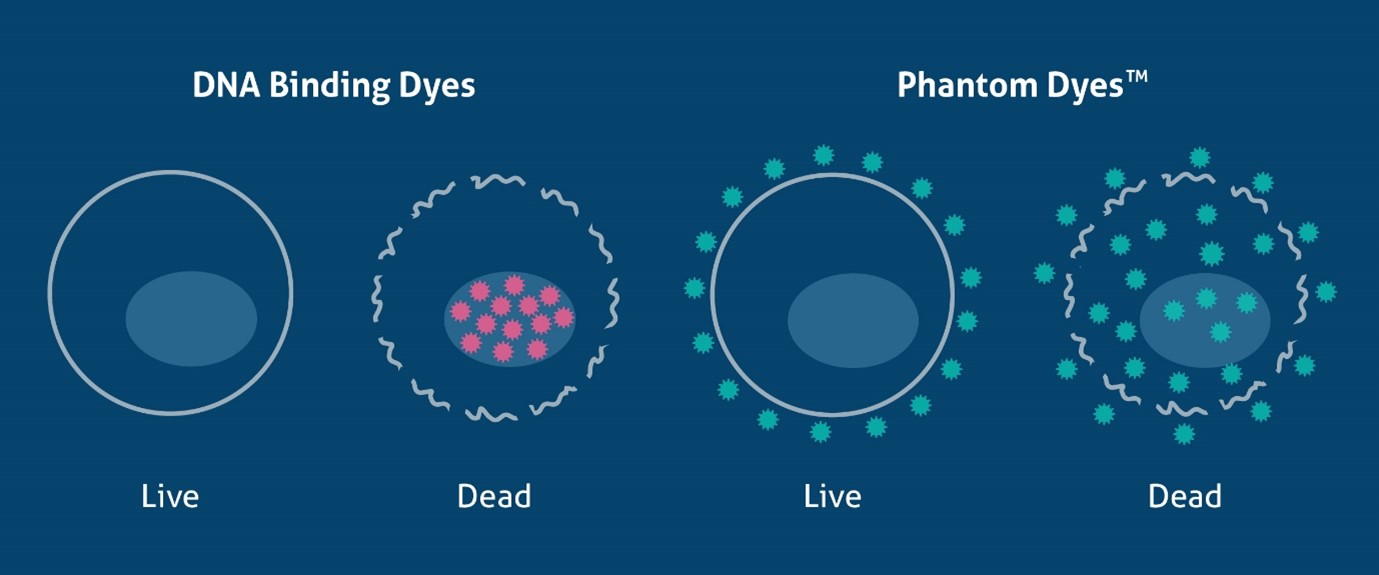
Figure 1. Schematic of the mechanism of DNA binding dyes and Phantom Dyes (fixable viability dyes), demonstrating how they differentiate live and dead cells.
Proteintech has the following viability dyes: Phantom Dyes:
PD00001 Phantom Dye Red 710 Viability Dye
PD00002 Phantom Dye Red 780 Viability Dye
PD00003 Phantom Dye UV 450 Viability Dye
PD00004 Phantom Dye Violet 450 Viability Dye
PD00005 Phantom Dye Violet 510 Viability Dye
PD00006 Phantom Dye Violet 540 Viability Dye
PD00007 Phantom Dye Blue 516 Viability Dye
Cell health assays
An alternative to fixable viability dyes is cell health assay kits, which use a combination of a viability dye and a DNA binding dye to determine the proportion of live and dead cells in your sample. These are commonly used in fluorescent live cell imaging as well as in flow cytometry.
One such viability dye is Calcein AM. Calcein AM is a non-fluorescent cell-permeant dye. Unlike the DNA binding dyes, this molecule can enter living cells. Once inside the cell, it undergoes acetoxymethyl ester hydrolysis by intracellular esterases, causing it to fluoresce in the green spectrum (excitation/emission: 495/520 nm). In contrast, dead cells have low esterase activity, so there is no fluorescence. This can then be paired with DNA binding dyes that fluoresce in different spectra such as PI (excitation/emission: 488/617 nm) or EthD-I (excitation/emission: 528/617 nm), to measure the proportion of dead cells and enabling the assessment of viability and toxicity in one assay (Figure 2). Of note, these assays do not work in cells that have been fixed as the dyes will not bind to any cellular components post-fixation, therefore read out of results should be done as soon as possible following staining.
Proteintech has a Viability/Cytotoxicity Assay Kit for Animal Live & Dead Cells (Calcein AM, PI Method, Cat# PF00007) and (Calcein AM, EthD-I Method, Cat# PF00008).
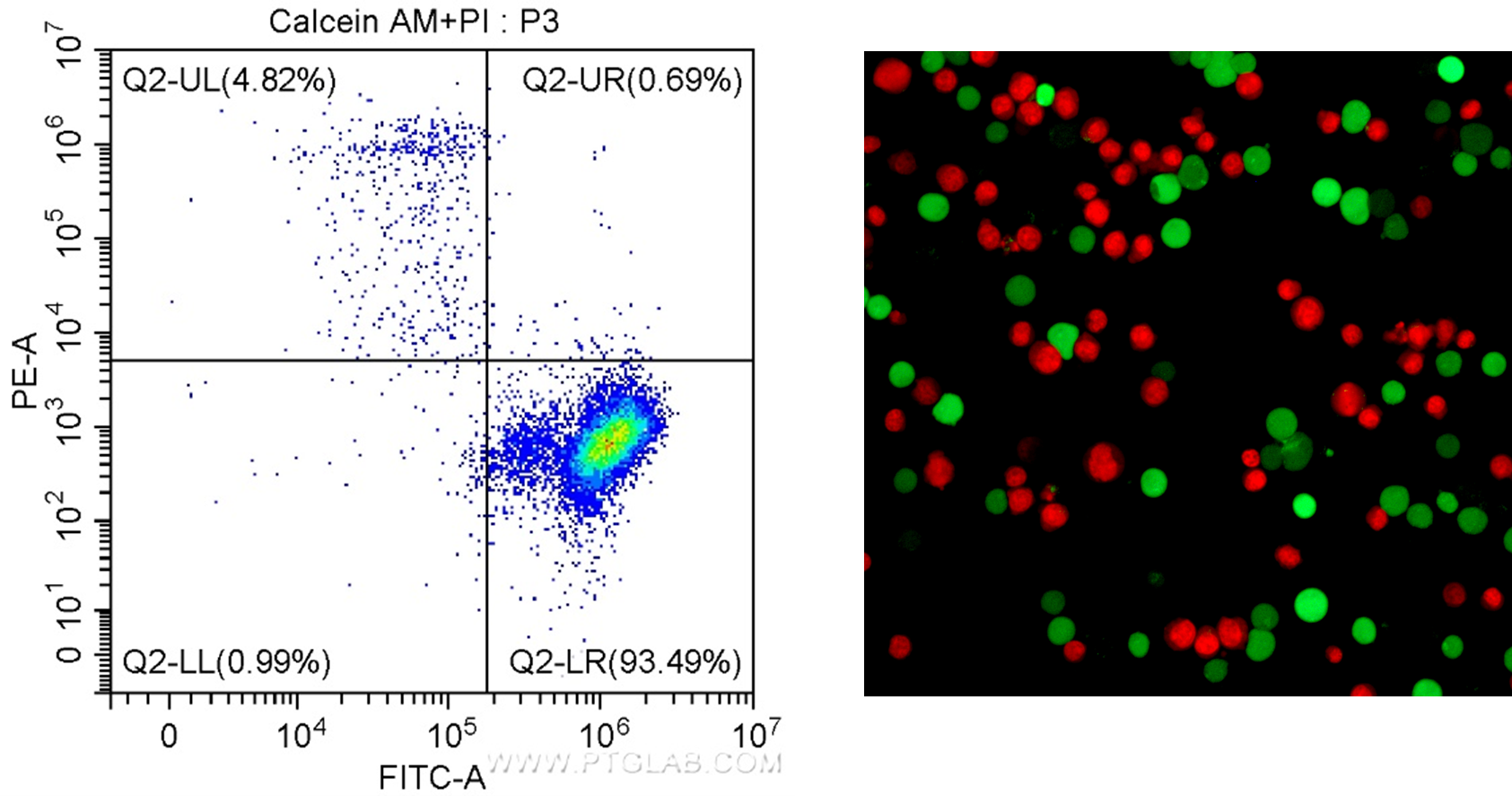
Figure 2. Results for the Viability/Cytotoxicity Assay Kit for Live & Dead Cells (Calcein AM, PI) Cat. # PF00007. Left: In flow cytometry, live cells have positive signal for Calcein AM (FITC/green detection), and dead cells can be visualized as positive for PI (PE-A/red detection). Right: Fluorescence microscopy of Jurkat cells stained with Calcein AM staining shows live cells in green, and PI staining shows dead cells in red.
Detecting apoptosis with Flow Cytometry
Apoptosis is a type of highly regulated cell death that is critical to development and maintaining homeostasis. Dysregulation can lead to diseases such as cancer and Alzheimer’s disease. More information can be found in our popular blog on apoptosis here.
Flow cytometry can be a useful tool in studying apoptosis, and there are many kits and dye combinations available for investigating this process. The most popular are Annexin V and TUNEL methods.
Annexin V binds to extracellular phosphatidylserine (PS), an early marker of apoptosis. Living cells contain PS on their inner, intracellular-facing side of the cell membrane. However, during the early stages of apoptosis, PS gets flipped to the extracellular side of the membrane. With PS exposed to the extracellular space, Annexin V binds PS and, combined with a fluorophore, generates a fluorescent signal to distinguish living cells from apoptotic cells. Proteintech’s CoraLite®488-Annexin V and PI apoptosis detection kit (PF00005) enables the observation of early and late apoptotic cells using fluorescently conjugated Annexin V in combination with PI (Figure 3).
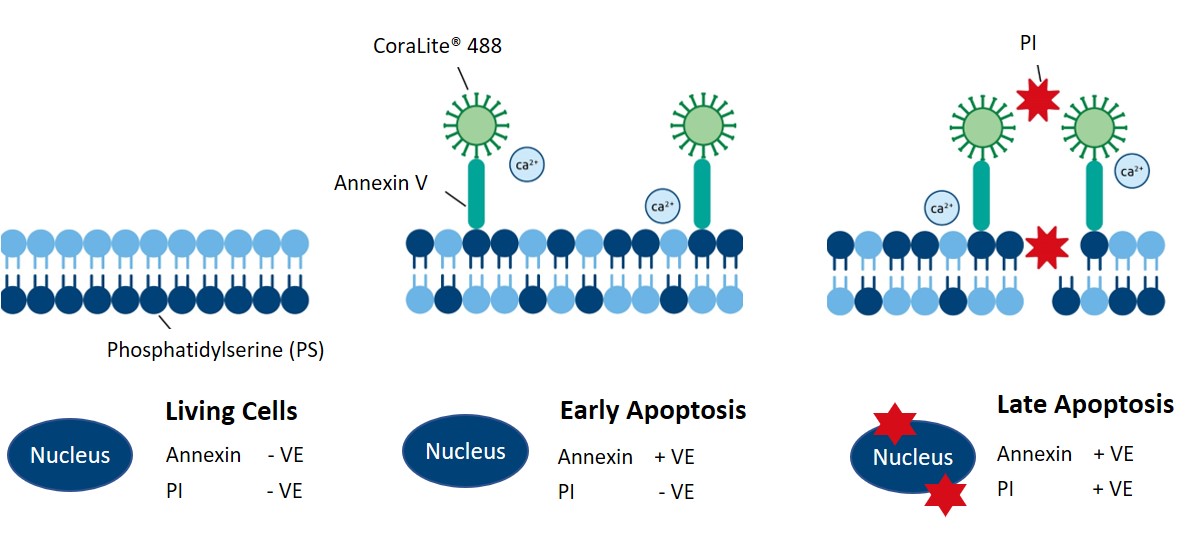
Figure 3. Schematic showing how Proteintech’s CoraLite®488-Annexin V and PI apoptosis detection kit labels early and late apoptotic cells. Catalog # PF00005.
Another popular way of detecting apoptosis is the TUNEL method. A hallmark of apoptosis is the degradation of cell chromosomal DNA. This process occurs in a specific and regular manner with DNA fragments occurring at different lengths in multiples of 180-200 base pairs, manifesting as a ladder-like staining pattern in agarose gel electrophoresis. The TUNEL method uses Terminal Deoxynucleotidyl Transferase (TdT) to catalyze the incorporation of fluorescently labelled dUTP into the 3'-OH end of the broken DNA of apoptotic cells. Labelled dUTP-DNA can then be directly observed with a fluorescence microscope or quantified by flow cytometry. Proteintech’s CoraLite®488 and CoraLite®594 TUNEL apoptosis detection kits (PF00006 and PF00009) allow the sensitive and selective detection of apoptotic dead cells, excluding dead necrotic cells or cells with DNA strand breaks.


